Laboratories
- Back
- Top > Laboratories > Oncology(Cooperating field) > Molecular Oncology
Oncology(Cooperating field)Molecular Oncology
Introduction
The mission of our laboratory is to elucidate the function and mechanism of genome and RNA and to apply them to better understand the molecular mechanisms of cancer and to develop the new approaches to cancer therapy. How do genes work? This fundamental question is crucial to understand the mechanisms of diverse diseases including cancer, which involve the interactions between genetic factors and environmental factors. Through an integrative omics analysis based on next-generation sequencing, bioinformatics, and genome/RNA engineering, we aim to get a deeper understanding of the relationships between genome, epigenome, transcription, and RNA to uncover the operating principles of gene regulation and disease mechanisms. We also aim to facilitate cancer precision medicine based on comprehensive omics analysis and develop new methodologies and technologies to advance this.
Research Projects
1. MicroRNA and Cancer
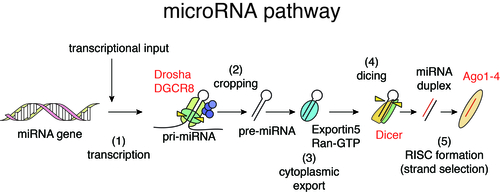
Function of many genes is governed by the flow of genetic information from DNA to RNA to protein, the central dogma of molecular biology. On the other hand, non-coding regions of the genome are pervasively transcribed and yield thousands of non-coding RNAs (ncRNAs). MicroRNAs (miRNAs) are abundant class of small ncRNAs with 21-25 nucleotides length. The first miRNA was discovered in 1993, and miRNA research was further accelerated by the Fire and Mello’s discovery of RNA interference (RNAi) in 1998.
In mammalian cells, the biogenesis of miRNAs is mediated by multiple biochemical steps including processing of miRNA precursors by two RNase III enzymes, Drosha and Dicer, and formation of RNA-induced silencing complex with Argonaute (Ago) proteins. miRNAs embedded in Ago proteins recognize their binding sequences predominantly located in 3’UTRs of target mRNAs through sequence complementarity between 5’ seed regions of miRNAs (nt 2-7) and their binding sites and suppress their expression. Reflecting the short sequence dependency, individual miRNAs regulate hundreds of target genes, thus forming complex gene regulatory networks. In addition to regulation of normal cells, miRNAs are known to modulate various pathological processes including cancer. We have investigated the relationships between cancer, miRNA biogenesis, and miRNA-mediated gene regulation and revealed the roles of tumor suppressor p53 in miRNA processing (Suzuki et al, Nature, 2009; Suzuki et al, Mol Cell, 2011).
In cancer biology, numerous studies have reported that miRNAs regulate many aspects of autonomous behavior of cancer cells, including sustained proliferation, resistance to cell death, and acquisition of metastasis phenotypes. We also previously reported that miRNAs regulate immune phenotypes of malignant lymphoma cells and modulate surrounding tumor microenvironments, illuminating miRNA-mediated non-cell-autonomous mechanism in cancer progression (Matsuyama et al, Blood, 2011; Suzuki et al, Oncogene, 2015).
2. Systems RNA Biology
Gene regulation by miRNAs is complicated with targeting of multiple mRNAs by individual miRNAs and their generally mild effects on target mRNAs. Understanding of such complex systems requires sophisticated approaches integrating multiple genome-wide datasets including transcriptomes. We have previously developed analytical approaches to infer miRNA activities from mRNA expression datasets and demonstrated that applying this approaches to analysis of paired miRNA-mRNA datasets improves identification of robust bio-markers in cancer (Suzuki et al, Nucleic Acid Res, 2013; Suzuki et al, Leukemia, 2013).
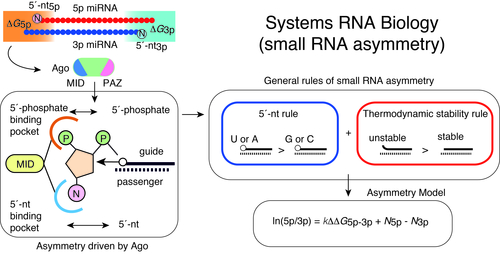
Of the two strands of miRNA duplexes, one miRNA strand is incorporated into an Ago protein, while the other strand is degraded. Asymmetric selection of one guide strand is crucial in miRNA-mediated gene regulation and specificity of RNAi. By combining molecular biology, biophysics, structural biology, and bioinformatics, we have presented a molecular and theoretical framework that describes the general features and underlying mechanisms of miRNA asymmetry and further presents modeling to estimate the asymmetry (Suzuki et al, Nat Struct Mol Biol, 2015; Matsuyama et al, IJMS, 2019). In consistent with the prediction by this model, cancer-associated miRNA variations were shown to modulate asymmetric selection. This model has contributed to development of synthetic RNA-Based immunomodulatory gene circuits for cancer immunotherapy (Nissim et al, Cell, 2017) and discovery of the first case of a pathogenic gain-of-function miRNA mutation in human diseases (Grigelioniene et al, Nature Medicine, 2019).
Combinatorial gene regulation by multiple miRNAs is widespread and closely spaced target sites often act cooperatively to achieve stronger repression (“neighborhood” miRNA cotargeting). Based on the functional studies of two miRNA alterations in disease pathogenesis, we have shown that miRNA cotargeting using extensively overlapping target sites (“seed overlap” miRNA cotargeting) increases susceptibility to downregulation by two miRNAs, consistent with additive but not cooperative recruitment of two miRNAs. Systematic characterization further revealed that extensive “seed overlap” is a prevalent feature among broadly conserved miRNAs. Intriguingly, highly conserved target sites of broadly conserved miRNAs are largely divided into two classes—those conserved among eutherian mammals and from human to Coelacanth, and the latter has a stronger association with both “seed overlap” and “neighborhood” miRNA cotargeting. These findings collectively suggests the complexity of distinct modes of miRNA cotargeting and the importance of their perturbations in human diseases (Kitai et al, BMC Biology, 2022).
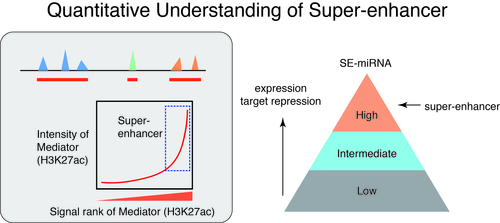
3. Biology of Transcription (1): Super-enhancer and Phase Separation
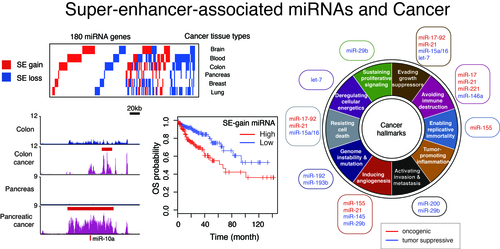
How do different cell types with the same genome establish cell-type-specific transcriptional programs? Cell-type-specific transcriptional programs are activated by the cell-type-specific enhancers bound by master transcription factors. Super-enhancers are emerging subclass of distal enhancers which regulate cell identity and disease processes. We have focused on the function of super-enhancers from the stand point of RNA. By integratively analyzing the relationships between super-enhancers and miRNAs, we have reported that super-enhancers have central roles in driving the function of cell-type-specific and evolutionally conserved miRNAs and maintaining their regulatory networks (Suzuki et al, Cell, 2017). In addition, we reported that super-enhancers facilitate processing of miRNA precursors, revealing unrecognized higher-order property of super-enhancers. Inspired from these findings, we currently instigate the roles of phase separation in gene regulation and enhancer control.
4. Biology of Transcription (1): Transcription Cycle
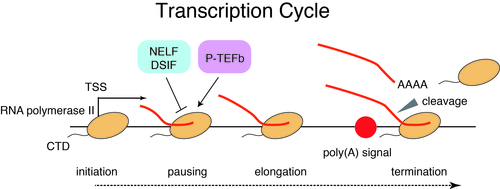
Toward better understanding of enhancer function, it is important to understand the dynamics of transcription across enhancers and the transcriptional impacts of enhancers on cognate promoters. Divergent transcription by RNA polymerase II occurs at many enhancers and promoters, yielding both coding mRNAs and diverse ncRNAs including eRNAs and uaRNAs. Furthermore, transcription by RNA polymerase II is regulated at multiple steps including initiation, pausing, pause release, elongation, and termination (“transcription cycle”). We have investigated the detailed mechanisms of transcription by integrating genome science and information science.
Protein-coding promoters are characterized by elongation of RNA polymerase II in the sense direction and early termination in the antisense direction. Such transcriptional directionality is modulated by asymmetrical distribution of U1 splicing motif and poly-adenylation signal (PAS motif) (U1-PAS axis). We have previously reported that premature PAS termination also frequently occurs in the sense direction and coincides with RNA polymerase II pause sites associated with +1 stable nucleosomes (Chiu et al, Mol Cell, 2018). Thus, promoter proximal Pol II pausing consists of two processes: transcription start site (TSS)-proximal pausing and +1 stable nucleosome pausing.
A recent advances in cancer genome studies have uncovered new class of cancer driver mutations affecting RNA processing and transcription. On the other hand, their functional significance has not been fully investigated in many cases. In myelodysplastic syndromes (MDS) and related neoplasms, the members of the cohesin complex including STAG2 are frequently mutated. In a recent collaboration study, we have importantly found that combined loss of STAG2 and master transcription factor RUNX1 synergistically attenuates enhancer–promoter loops, leading to development of MDS, and that cohesin deficiency preferentially downregulates genes with high basal transcriptional pausing, explaining the feature of selective gene dysregulation (Ochi et al, Cancer Discovery, 2020).
5. Multi-Omics Analysis in Cancer
We also aim to identify new molecular targets in cancer therapies through integrative omics analysis. In a previous study, by comparing super-enhancers in normal cells/tissues and cancer cells, we have reported that gain and loss of super-enhancers are frequently found in the neighborhoods of oncogenic and tumor-suppressive miRNAs associated with various cancer hallmark traits (Suzuki et al, Cell, 2017). Furthermore, we have recently reported the first case of miRNA gain-of-function mutation in human disease (Grigelioniene et al, Nature Medicine, 2019), and in a companion study, we performed systematic analysis of siRNA off-target data and genome-wide RNAi cancer lethality screens using 501 human cancer cell lines, a cancer dependency map, and offered a basis for improvement of RNAi-based functional genomics (Suzuki et al, Nature Genetics, 2018).
6. beyond
By expanding the expertise in computational modeling in previous studies, a recent collaboration project with Kyushu University has successfully developed new CRISPR-based genome editing strategy, called “safeguard sgRNA” strategy, which improves the safety and applicability of genome editing (Kawamata et al, Nature Biomedical Engineering, 2023).
Faculty Members
| Faculty | Position | Department |
|---|---|---|
| Hiroshi Suzuki | Professor | Molecular Oncology |
| Koichi Ogami | Assistant professor | Molecular Oncology |
| Seiko Yoshino | Designated Assistant Professor | Molecular Oncology |
| Shintaro Komatsu | Researcher | Molecular Oncology |
Bibliography
- 2023
- Kawamata M, Suzuki HI, Kimura R, Suzuki A. Optimization of Cas9 activity through the addition of cytosine extensions to single-guide RNAs. Nat Biomed Eng, 2023; 7(5): 672-691.
- Suzuki HI. Roles of MicroRNAs in Disease Biology. JMAJ, 2023; 6(2): 104-113.
- 2022
- Suzuki HI, Onimaru K. Biomolecular Condensates in Cancer Biology. Cancer Sci, 2022; 113(2): 382-391.
- Yoshino S, Suzuki HI. The molecular understanding of super-enhancer dysregulation in cancer. Nagoya J. Med. Sci, 2022; 84: 216-229.
- Miyakawa K, Miyashita N, Horie M, Terasaki Y, Tanaka H, Urushiyama H, Fukuda K, Okabe Y, Ishii T, Kuwahara N, Suzuki HI, Nagase T, Saito A. Cancer Sci, 2022; 113(11): 3932-3946.
- Shimamura Y, Furuhashi K, Tanaka A, Karasawa M, Nozaki T, Komatsu S, Watanabe K, Shimizu A, Minatoguchi S, Matsuyama M, Sawa Y, Tsuboi N, Ishimoto T, Suzuki HI, Maruyama S. Mesenchymal stem cells exert renoprotection via extracellular vesicle-mediated modulation of M2 macrophages and spleen-kidney network. Commun Biol, 2022; 5(1): 753.
- Yan M, Komatsu N, Muro R, Huynh NC, Tomofuji Y, Okada Y, Suzuki HI, Takaba H, Kitazawa R, Kitazawa S, Pluemsakunthai W, Mitsui Y, Satoh T, Okamura T, Nitta T, Im SH, Kim CJ, Kollias G, Tanaka S, Okamoto K, Tsukasaki M, Takayanagi H. ETS1 governs pathological tissue-remodeling programs in disease-associated fibroblasts. Nat Immunol, 2022; 23(9): 1330-1341.
- Kitai H, Kato N, Ogami K, Komatsu S, Watanabe Y, Yoshino S, Koshi E, Tsubota S, Funahashi Y, Maeda T, Furuhashi K, Ishimoto T, Kosugi T, Maruyama S, Kadomatsu K, Suzuki HI. Systematic characterization of seed overlap microRNA cotargeting associated with lupus pathogenesis. BMC Biol, 2022; 20(1): 248.
- 2021
- Ogami K, Suzuki HI. Nuclear RNA Exosome and Pervasive Transcription: Dual Sculptors of Genome Function. Int J Mol Sci, 2021; 22(24): 13401.
- 2020
- Miyashita N, Horie M, Suzuki HI, Saito M, Mikami Y, Okuda K, Boucher RC, Suzukawa M, Hebisawa A, Saito A, Nagase T. FOXL1 Regulates Lung Fibroblast Function via Multiple Mechanisms. Am J Respir Cell Mol Biol. 2020; 63(6): 831-842.
- Li Y, Que L, Fukano K, Koura M, Kitamura K, Zheng X, Kato T, Aly HH, Watashi K, Tsukuda S, Aizaki H, Watanabe N, Sato Y, Suzuki T, Suzuki HI, Hosomichi K, Kurachi M, Wakae K, Muramatsu M. MCPIP1 reduces HBV-RNA by targeting its epsilon structure. Sci Rep. 2020; 10(1): 20763.
- Tomofuji Y, Takaba H, Suzuki HI, Benlaribi R, Martinez CDP, Abe Y, Morishita Y, Okamura T, Taguchi A, Kodama T, Takayanagi H. Chd4 choreographs self-antigen expression for central immune tolerance. Nat Immunol. 2020; 21(8): 892-901.
- Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, Kataoka K, Koseki H, Nakayama M, Morishita D, Tsuruyama T, Saiki R, Yoda A, Okuda R, Yoshizato T, Yoshida K, Shiozawa Y, Nannya Y, Kotani S, Kogure Y, Kakiuchi N, Nishimura T, Makishima H, Malcovati L, Yokoyama A, Takeuchi K, Sugihara E, Sato TA, Sanada M, Takaori-Kondo A, Cazzola M, Kengaku M, Miyano S, Shirahige K, Suzuki HI, Ogawa S. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov, 2020; 10(6): 836-853.
- 2019
- Matsuyama H, Suzuki HI. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int J Mol Sci, 2019; 21(1): 132.
- Tominaga K, Suzuki HI. TGF-β Signaling in Cellular Senescence and Aging-Related Pathology. Int J Mol Sci, 2019; 20(20): 5002.
- Kelly TJ, Suzuki HI, Zamudio JR, Suzuki M, Sharp PA. Sequestration of microRNA-mediated target repression by the Ago2-associated RNA-binding protein FAM120A. RNA, 2019; 25(10): 1291-1297.
- Akatsu Y, Takahashi N, Yoshimatsu Y, Kimuro S, Muramatsu T, Katsura A, Maishi N, Suzuki HI, Inazawa J, Hida K, Miyazono K, Watabe T. Fibroblast growth factor signals regulate transforming growth factor-β-induced endothelial-to-myofibroblast transition of tumor endothelial cells via Elk1. Mol Oncol, 2019; 13(8): 1706-1724.
- Grigelioniene G, Suzuki HI, Taylan F, Mirzamohammadi F, Borochowitz ZU, Ayturk UM, Tzur S, Horemuzova E, Lindstrand A, Weis MA, Grigelionis G, Hammarsjö A, Marsk E, Nordgren A, Nordenskjöld M, Eyre DR, Warman ML, Nishimura G, Sharp PA, Kobayashi T. Gain-of-function mutation of microRNA-140 in human skeletal dysplasia. Nat Med, 2019; 25(4): 583-590.
- 2018
- Souma K, Shichino S, Hashimoto S, Ueha S, Tsukui T, Nakajima T, Suzuki HI, Shand FHW, Inagaki Y, Nagase T, Matsushima K. Lung fibroblasts express a miR-19a-19b-20a sub-cluster to suppress TGF-β-associated fibroblast activation in murine pulmonary fibrosis. Sci Rep, 2018; 8(1): 16642.
- Gao A, Shrinivas K, Lepeudry P, Suzuki HI, Sharp PA, Chakraborty AK. Evolution of weak cooperative interactions for biological specificity. Proc Natl Acad Sci U S A, 2018; 115(47): E11053-E11060.
- Suzuki HI, Horie M, Mihira H, Saito A. Molecular Analysis of Endothelial-mesenchymal Transition Induced by Transforming Growth Factor-β Signaling. J Vis Exp, 2018; (138): 57577.
- Miyashita N, Horie M, Suzuki HI, Yoshihara M, Djureinovic D, Persson J, Brunnström H, Lindskog C, Elfving H, Micke P, Saito A, Nagase T. An Integrative Analysis of Transcriptome and Epigenome Features of ASCL1-Positive Lung Adenocarcinomas. J Thorac Oncol, 2018; 13(11): 1676-1691.
- Suzuki HI. MicroRNA Control of TGF-β Signaling. Int J Mol Sci, 2019; 19(7): 1901.
- Suzuki HI, Spengler RM, Grigelioniene G, Kobayashi T, Sharp PA. Deconvolution of seed and RNA-binding protein crosstalk in RNAi-based functional genomics. Nat Genet, 2018; 50(5): 657-661.
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, Sharp PA. Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP. Mol Cell, 2018; 69(4): 648-663.
- Lai HH, Li JN, Wang MY, Huang HY, Croce CM, Sun HL, Lyu YJ, Kang JW, Chiu CF, Hung MC, Suzuki HI, Chen PS. HIF-1α promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J Clin Invest, 2018; 128(2): 625-643.
- Horie M, Miyashita N, Mikami Y, Noguchi S, Yamauchi Y, Suzukawa M, Fukami T, Ohta K, Asano Y, Sato S, Yamaguchi Y, Ohshima M, Suzuki HI, Saito A, Nagase T. TBX4 is involved in the super-enhancer-driven transcriptional programs underlying features specific to lung fibroblasts. Am J Physiol Lung Cell Mol Physiol, 2018; 314(1): L177-L191.
- 2017
- Nissim L, Wu MR, Pery E, Binder-Nissim A, Suzuki HI, Stupp D, Wehrspaun C, Tabach Y, Sharp PA, Lu TK. Synthetic RNA-Based Immunomodulatory Gene Circuits for Cancer Immunotherapy. Cell, 2017; 171(5): 1138-1150.
- Suzuki HI, Katsura A, Mihira H, Horie M, Saito A, Miyazono K. Regulation of TGF-β-mediated endothelial-mesenchymal transition by microRNA-27. J Biochem, 2017; 161(5): 417-420.
- Suzuki HI, Young RA, Sharp PA. Super-Enhancer-Mediated RNA Processing Revealed by Integrative MicroRNA Network Analysis. Cell, 2017; 168(6): 1000-1014.
- 2016
- Horie M, Saito A, Ohshima M, Suzuki HI, Nagase T. YAP and TAZ modulate cell phenotype in a subset of small cell lung cancer. Cancer Sci, 2016; 107(12): 1755-1766.
- Katsura A, Suzuki HI, Ueno T, Mihira H, Yamazaki T, Yasuda T, Watabe T, Mano H, Yamada Y, Miyazono K. MicroRNA-31 is a positive modulator of endothelial-mesenchymal transition and associated secretory phenotype induced by TGF-β. Genes Cells, 2016; 21(1): 99-116.
- 2015
- Shichino S, Abe J, Ueha S, Otsuji M, Tsukui T, Kosugi-Kanaya M, Shand FH, Hashimoto S, Suzuki HI, Morikawa T, Inagaki Y, Matsushima K. Reduced supply of monocyte-derived macrophages leads to a transition from nodular to diffuse lesions and tissue cell activation in silica-induced pulmonary fibrosis in mice. Am J Pathol, 2015; 185(11): 2923-2938.
- Sugimoto K, Suzuki HI, Fujimura T, Ono A, Kaga N, Isobe Y, Sasaki M, Taka H, Miyazono K, Komatsu N. A clinically attainable dose of L-asparaginase targets glutamine addiction in lymphoid cell lines. Cancer Sci, 2015; 106(11): 1534-1543.
- Suzuki HI, Katsura A, Yasuda T, Ueno T, Mano H, Sugimoto K, Miyazono K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat Struct Mol Biol, 2015; 22(7): 512-521.
- Suzuki HI, Katsura A, Miyazono K. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci, 2015; 106(9): 1174-1181.
- Itami S, Eguchi Y, Mizutani T, Aoki E, Ohgi T, Kuroda M, Ochiya T, Kato N, Suzuki HI, Kawada N, Murakami Y. Control of HCV Replication With iMIRs, a Novel Anti-RNAi Agent. Mol Ther Nucleic Acids, 2015; 4(1): e219.
- Suzuki HI, Katsura A, Matsuyama H, Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene, 2015; 34(24): 3085-3094.
- 2014
- Horie M, Saito A, Noguchi S, Yamaguchi Y, Ohshima M, Morishita Y, Suzuki HI, Kohyama T, Nagase T. Differential knockdown of TGF-β ligands in a three-dimensional co-culture tumor- stromal interaction model of lung cancer. BMC Cancer, 2014; 14: 580.
- Noguchi S, Saito A, Horie M, Mikami Y, Suzuki HI, Morishita Y, Ohshima M, Abiko Y, Mattsson JS, König H, Lohr M, Edlund K, Botling J, Micke P, Nagase T. An integrative analysis of the tumorigenic role of TAZ in human non-small cell lung cancer. Clin Cancer Res, 2014; 20(17): 4660-4672.
- Tanaka M, Suzuki HI, Shibahara J, Kunita A, Isagawa T, Yoshimi A, Kurokawa M, Miyazono K, Aburatani H, Ishikawa S, Fukayama M. EVI1 oncogene promotes KRAS pathway through suppression of microRNA-96 in pancreatic carcinogenesis. Oncogene, 2014; 33(19): 2454-2463.
- 2013
- Yoshimatsu Y, Lee YG, Akatsu Y, Taguchi L, Suzuki HI, Cunha SI, Maruyama K, Suzuki Y, Yamazaki T, Katsura A, Oh SP, Zimmers TA, Lee SJ, Pietras K, Koh GY, Miyazono K, Watabe T. Bone morphogenetic protein-9 inhibits lymphatic vessel formation via activin receptor-like kinase 1 during development and cancer progression. Proc Natl Acad Sci U S A, 2013; 110(47): 18940-18945.
- Suzuki HI, Matsuyama H, Noguchi M, Yao T, Komatsu N, Mano H, Sugimoto K, Miyazono K. Computational dissection of distinct microRNA activity signatures associated with peripheral T cell lymphoma subtypes. Leukemia, 2013; 27(10): 2107-2111.
- Saito A, Suzuki HI, Horie M, Ohshima M, Morishita Y, Abiko Y, Nagase T. An integrated expression profiling reveals target genes of TGF-β and TNF-α possibly mediated by microRNAs in lung cancer cells. PLoS One, 2013; 8(2): e56587.
- Suzuki HI, Mihira H, Watabe T, Sugimoto K, Miyazono K. Widespread inference of weighted microRNA-mediated gene regulation in cancer transcriptome analysis. Nucleic Acids Res, 2013; 41(5): e62.
- Suzuki HI, Miyazono K. p53 actions on microRNA expression and maturation pathway. Methods Mol Biol, 2013; 962: 165-181.
- Liang C, Xiong K, Szulwach KE, Zhang Y, Wang Z, Peng J, Fu M, Jin P, Suzuki HI, Liu Q.. Sjogren syndrome antigen B (SSB)/La promotes global microRNA expression by binding microRNA precursors through stem-loop recognition. J Biol Chem, 2013; 288(1): 723-736.
- 2012
- Suzuki HI, Miyazono K. Control of MicroRNA Maturation by p53 Tumor Suppressor and MCPIP1 Ribonuclease. The Enzymes, 2012; 32: 163-183.
- Nishimori H, Ehata S, Suzuki HI, Katsuno Y, Miyazono K. Prostate cancer cells and bone stromal cells mutually interact with each other through bone morphogenetic protein-mediated signals. J Biol Chem, 2012; 287(24): 20037-20046.
- Mihira H, Suzuki HI, Akatsu Y, Yoshimatsu Y, Igarashi T, Miyazono K, Watabe T. TGF-β-induced mesenchymal transition of MS-1 endothelial cells requires Smad-dependent cooperative activation of Rho signals and MRTF-A. J Biochem, 2012; 151(2): 145-156.
- 2011
- Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell, 2011; 44(3): 424-436.
- Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N, Mano H, Sugimoto K, Miyazono K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood, 2011; 118(26): 6881-6892.
- Mizutani A, Koinuma D, Tsutsumi S, Kamimura N, Morikawa M, Suzuki HI, Imamura T, Miyazono K, Aburatani H. Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4alpha in HepG2 cells. J Biol Chem, 2011; 286(34): 29848-29860.
- Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem, 2011; 149(1): 15-25.
- 2010
- Suzuki HI, Miyazono K. Dynamics of microRNA biogenesis: crosstalk between p53 network and microRNA processing pathway. J Mol Med (Berl), 2010; 88(11): 1085-1094.
- Suzuki HI, Kiyono K, Miyazono K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy, 2010; 6(5): 645-647.
- Hosoi M, Nannya Y, Sasaki T, Suzuki HI, Ueda K, Tsujino T, Isayama H, Takahashi T, Koike K, Kurokawa M. Biliary cast syndrome and benign biliary stricture as complications of allogeneic hematopoietic stem cell transplantation. Ann Hematol, 2010; 89(12): 1287-1289.
- Suzuki HI, Hosoya N, Miyagawa K, Ota S, Nakashima H, Makita N, Kurokawa M. Erdheim-Chester disease: multisystem involvement and management with interferon-alpha. Leuk Res, 2010; 34(1): e21-e24.
- 2009
- Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, Sugimoto K, Miyazono K. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res, 2009; 69(23): 8844-8852.
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature, 2009; 460(7254): 529-533.
- Kiyono K, Suzuki HI, Morishita Y, Komuro A, Iwata C, Yashiro M, Hirakawa K, Kano MR, Miyazono K. c-Ski overexpression promotes tumor growth and angiogenesis through inhibition of transforming growth factor-beta signaling in diffuse-type gastric carcinoma. Cancer Sci, 2009; 100(10): 1809-1816.
- Komuro A, Yashiro M, Iwata C, Morishita Y, Johansson E, Matsumoto Y, Watanabe A, Aburatani H, Miyoshi H, Kiyono K, Shirai YT, Suzuki HI, Hirakawa K, Kano MR, Miyazono K. Diffuse-type gastric carcinoma: progression, angiogenesis, and transforming growth factor beta signaling. J Natl Cancer Inst, 2009; 101(8): 592-604.
- 2002
- Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature, 2002; 416(6882): 744-749.
- 2008
- Suzuki H, Tsurita G, Ishihara S, Akahane M, Kitayama J, Nagawa H. Resovist-enhanced MRI for preoperative assessment of colorectal hepatic metastases: a case of multiple bile duct hamartomas associated with colon cancer. Case Rep Gastroenterol, 2008; 2(3): 509-516.
- Suzuki HI, Izutsu K, Watanabe T, Oshima K, Kanda Y, Motokura T, Chiba S, Kurokawa M. Late-onset pneumatosis cystoides intestinalis associated with non-infectious pulmonary complications after allogeneic hematopoietic stem cell transplantation. Int J Hematol, 2008; 88(1): 116-118.
- Suzuki HI, Suzuki T, Kamijo A, Oota S, Sato H, Hangaishi A, Takahashi T, Kanda Y, Motokura T, Chiba S, Kurokawa M. Antileukemic immunity associated with antineutrophil antibody production after allogeneic hematopoietic SCT for myeloid/NK-cell precursor acute leukemia. Bone Marrow Transplant, 2008; 42(4): 285-287.
- Suzuki HI, Hangaishi A, Hosoya N, Watanabe T, Kanda Y, Motokura T, Chiba S, Kurokawa M. Herpes simplex encephalitis and subsequent cytomegalovirus encephalitis after chemoradiotherapy for central nervous system lymphoma: a case report and literature review. Int J Hematol, 2008; 87(5): 538-541.
- Suzuki HI, Asai T, Tamaki Z, Hangaishi A, Chiba S, Kurokawa M. Drug-induced hypersensitivity syndrome with rapid hematopoietic reconstitution during treatment for acute myeloid leukemia. Haematologica, 2008; 93(3): 469-470.
- Oka M, Iwata C, Suzuki HI, Kiyono K, Morishita Y, Watabe T, Komuro A, Kano MR, Miyazono K. Inhibition of endogenous TGF-beta signaling enhances lymphangiogenesis. Blood, 2008; 111(9): 4571-4579.
- Suzuki HI, Asai T, Okada K, Kazuyama Y, Takahashi T, Kanda Y, Chiba S, Kurokawa M. Disseminated adenovirus disease by multiple adenovirus serotypes following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant, 2008; 14(3): 353-355.
- 2007
- Ogawa K, Saito A, Matsui H, Suzuki H, Ohtsuka S, Shimosato D, Morishita Y, Watabe T, Niwa H, Miyazono K. Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells. J Cell Sci, 2007; 120: 55-65.
Research Keywords
cancer, genome, RNA, microRNA, non-coding RNA, transcription, epigenome, enhancer, super-enhancer, phase separation, next-generation sequencer, bioinformatics, genome engineering, gene screening, single cell analysis
Our website has been renewed. (2023.8)
Invitation
We are recruiting staffs and graduate students from all over the world. Please send your curriculum vitae, bibliography, and a letter describing your research or clinical experience by e-mail to us.
Graduate students can be supported by several fellowship programs including CIBoG, Nagoya University Interdisciplinary Frontier Fellowship, and THERS Interdisciplinary Frontier Next Generation Researcher.
Access
Hiroshi Suzuki, M.D., Ph.D.
Professor
Division of Molecular Oncology
Center for Neurological Diseases and Cancer
Nagoya University Graduate School of Medicine
65 Tsurumai-cho, Showa-ku, Nagoya, 466-8550, Japan
E-mail: hisuzuki@med.nagoya-u.ac.jp