Laboratories
- Back
- Top > Laboratories > Laboratory Animal Science(Cooperating field) > Laboratory Animal Science
Laboratory Animal Science(Cooperating field)Laboratory Animal Science
Introduction
In humans, genetic analysis of complex traits is made very difficult by population heterogeneity, environmental factors, and other considerations. In contrast, a genetic approach in the mouse represents an important tool by utilizing many powerful techniques such as the availability of numerous well-defined inbred strains and genetic manipulation (TG, KO, KI). One approach to identify host resistance genes is positional cloning or positional candidate methods that are based on the forward genetics in mice. Our study may provide valuable insights into the molecular basis of human disease.
Research Projects
- Systematic genetic analysis for disease susceptibility gene in mice
SM/J and A/J mice possess many different phenotypic traits such as pulmonary adenoma, body weight, blood lipid concentrations, type 2 diabetes, susceptibility to nematode infection and circadian rhythms. The chromosomes in each recombinant inbred (RI) strain are a genetically unique mixture of those of the parental strains. A set of SMXA RI strains derived from SM/J and A/J mice provides a valuable resource for the chromosomal mapping of complex genetic traits. Consomic strains in which one chromosome is derived from one strain and the remainder are derived from another. We established consomic strains derived by using A/J as the recipient strain and SM/J as the donor strain. Combination of previously established set of SMXA RI strains and these consomic strains (Fig.1) will provide a powerful resource for systematic analysis of complex genetic traits in mice.
Fig.1 Schema of chromosome construction of systematic genetic analysis tool in mice - Genetic analysis for host resistance gene to parasitic infection in mice
Parasitic diseasesuch as malaria (Fig.2) affects many people and results in more than 1 million deaths annually. Host genetic factors have been established to influence the outcome of this disease. Experimental models in mice are very useful for revealing complex host genetic factors of human parasitic disease. Our approach to identify host resistance genes is positional cloning or positional candidate methods that are based on the forward genetics in mice. Our study may provide valuable insights into the molecular basis of host resistance to parasite infections, which could have important implications for the study of host defense in humans.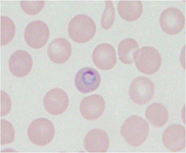
Fig.2 rodent malaria parasite in RBC of mice
Faculty Members
| Faculty | Position | Department |
|---|---|---|
| Tamio Ohno | Associate Professor | Laboratory Animal Science |
| Yuki Miyasaka | Assistant Professor | Laboratory Animal Science |
Bibliography
- 2016
- Yashima A, Mizuno M, Yuzawa Y, Shimada K, Suzuki N, Tawada H, Sato W, Tsuboi N, Maruyama S, Ito Y, Matsuo S, Ohno T: Mesangial proliferative glomerulonephritis in murinemalaria parasite, Plasmodium chabaudi AS, infected NC mice. Clin Exp Nephrol, in press.
- Miyasaka Y, Shitara H, Suzuki S, Yoshimoto S, Seki Y, Ohshiba Y, Okumura K, Taya C, Tokano H, Kitamura K, Takada T, Hibino H, Shiroishi T, Kominami R, Yonekawa H, Kikkawa Y. Heterozygous mutation of Ush1g/Sans in mice causes early-onset progressive hearing loss, which is recovered by reconstituting the strain-specific mutation in Cdh23. Hum Mol Genet, 2016; 25: 2045-2059.
- Suzuki M, Kobayashi M, Ohno T, Kanamori S, Tateishi S, Murai A, Horio F. Genetic dissection of the fatty Liver QTL Fl1sa by using congenic mice and identification of candidate genes in the liver and epididymal fat. BMC Genet, 2016; 17:145.
- Kobayashi M, Suzuki M, Ohno T, Tsuzuki K, Taguchi C, Tateishi S, Kawada T, Kim YI, Murai A, Horio F. Detection of differentially expressed candidate genes for a fatty liver QTL on mouse chromosome 12. BMC Genet, 2016; 17: 73.
- 2014
- Kobayashi M, Ohno T, Ihara K, Murai A, Kumazawa M, Hoshino H, Iwanaga K, Iwai H, Hamana Y, Ito M, Ohno K, Horio F. Searching for genomic region of high-fat diet-induced type 2 diabetes in mouse chromosome 2 by analysis of congenic strains. PLoS One, 2014; 9: e96271.
- 2013
- Ohno T, Okamoto M, Hara T,Hashimoto N, Imaizumi K, Matsushima M, Nishimura M, Shimokata K, Hasegawa Y, Kawabe T. Detection of loci for allergic asthma using SMXA recombinant inbred strains of mice. Immunogenetics, 2013; 65:17-24.
- 2012
- Ohno T, Hata K, Baba T, Io F, Kobayashi M, Horio F, Nishimura M. Establishment of consomic strains derived from A/J and SM/J mice for genetic analysis of complex traits. Mamm Genome, 2012; 23: 764-769.
Research Keywords
mouse, disease susceptibility gene, forward genetics, genome editing, parasitic disease, malaria, metabolic disorder