Laboratories
- Back
- Top > Laboratories > Biomedical Regulation > Infectious Diseases
Biomedical RegulationInfectious Diseases
Introduction
Our research team consists of 1 professor, 3 assistant professor, 4 graduate students (including two students working at other hospital and educational facility), and two young medical staffs. Our main research projects are first, control of drug resistant bacteria, or healthcare associated infections, second, elucidation of mechanism of diseases caused by non-tuberculous mycobacteria (NTM).
We reported the results of point prevalence survey in Japan for prevention and control of healthcare associated infections for the first time. Risk factors for and mechanism of resistance of carbapenems in P. aeruginosa, molecular epidemiological and genetic characterization of M. avium subsp. hominissuis and M. intracellulare isolates from different geographic regions, of M. avium subsp. hominissuis isolates from humans and swines were the themes of our research. Moreover, we are now working on the control of antimicrobial resistance including CRE which is becoming significant threat in the crinical settings, and ESBL-producing Klebsiella.
Research Projects
1.Prevention and control for healthcare associated infections and antimicrobial resistance.
i) Point prevalence survey
Pilot PPS study was conducted to evaluate the epidemiology of healthcare–associated infections (HAIs) and antimicrobial use in a Japanese tertiary university hospital. A 1-day, cross-sectional PPS revealed that of 841 patients, 85 (10.1%) had 90 active HAIs, and 308 patients (36.6%) were administered some antimicrobials.The frequent infections were pneumonia (20.0%) and surgical site infections (16.7%). The most common pathogen isolated were Enterobacteriaceae (27.6%). Of the 118 antimicrobials used for treatment of HAIs, carbapenems were the most frequently administered category of antimicrobials (22.9%). In regard to antimicrobials for surgical prophylaxis, 37 of 119 (31.1%) were administered to patients on postoperative day 3 or later, and 48 of 119 (40.3%) were administered orally. This first PPS in Japan showed that incidence of HAIs is higher than in other developed countries. The social and medical situations in Japan may affect patient demographics, active HAIs, and antimicrobial use.
In 2016, we conducted multicenter PPS in collaboration with Kyoto Univ., Hiroshima Univ., and Nara Prefectural Medical Univ.
ii) Carbapenem resistance in Pseudomonas aeruginosa
Clinical isolates of P. aeruginosa whose MICs of imipenem or meropenem showed 4-8mg/ml were frequently encountered in clinical settings. Multiple logistic regression analysis showed that clinical risk factors for the ≧4-fold increase in MICs of carbapenems were prior use of carbapenems (OR, 2.799; 95% CI, 1.088-7.200, p=0.033) and use of ventilator or tracheostomy (OR, 2.648; 95% CI, 1.051-6.671, p=0.039). Nineteen pairs of genetically identical P. aeruginosa isolates from the same patient, which showed more than or equal to 4-fold increase in MICs of carbapenems were used for the analysis of oprD gene and the expression of oprD protein. Loss of functional OprD protein due to the mutations of oprD gene tended to occur in P. aeruginosa isolates whose MIC of IPM was more than 8 mg/mL, and the reduction of OprD expression occurred in isolates whose MIC of IPM was 4 or 8 μg/mL.
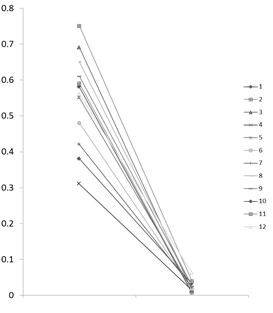
OprD expression IPM-MIC≥8μg/ml
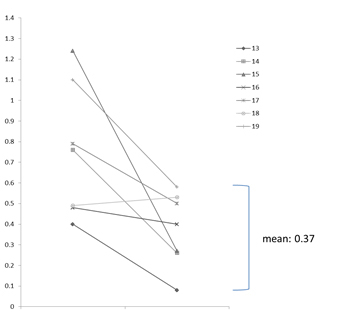
OprD expression IPM-MIC=4 or 8μg/ml
iii) Prevention and control of carbapenem resistant Enterobacteriaceae
The spread of CRE is now global threat, taking us back to a pre-antibiotic era. In Japan, the national surveillance has started for cases infected by CRE. In our laboratory, we are working on an effective detection method, infection control measures, and therapy, analyses on mechanisms of resistance, molecular epidemiology of the CRE clinical isolates.
iv) ESBL-producing Klebsiella
Compared with E. coli, ESBL-producing Klebsiella is not now prevailing, and no high-risk clone was not still found. We are now collaborating with Kyushu Univ. and Asahikawa Medical School to reveal the molecular epidemiology and genetic background of ESBL-producing Klebsiella spp.
2. Elucidation of NTM diseases from bacterial side
i) Genetic diversity of clinical Mycobacterium avium subsp. hominissuis and
Mycobacterium intracellulare isolates causing pulmonary diseases.
Mycobacterium avium complex (MAC) infections are increasing annually inmany countries. MAC strains are the most common nontuberculous mycobacterial pathogens isolated from respiratory samples and predominantly consist of two species, Mycobacterium avium and Mycobacterium intracellulare. We analyzed the molecular epidemiology and genetic backgrounds of clinical MAC isolates collected from The
Netherlands, Germany, United States, Korea and Japan. Variable numbers of tandem repeats (VNTR) analysis was used to examine the genetic relatedness of clinical isolates of M. avium subsp. hominissuis (n = 261) and M. intracellulare (n=116). Minimum spanning tree and unweighted pair group method using arithmetic averages analyses based on the VNTR data indicated that M. avium subsp. hominissuis isolates from Japan shared a high degree of genetic relatedness with Korean isolates, but not with isolates from Europe or the United States, whereas M. intracellulare isolates did not show any specific clustering by geographic origin.
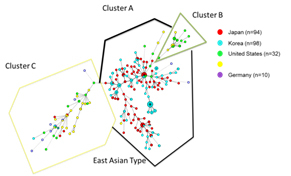
M. avium subsp. hominissuis VNTR
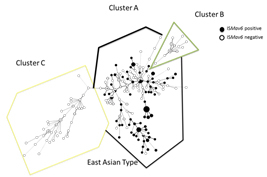
M. avium subsp. hominissuis ISMav6
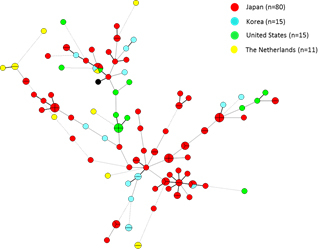
M. intracellulare VNTR
ii) Molecular typing and genetic characterization of Mycobacterium avium subspecies hominissuis isolates from humans and swine.
Mycobacterium avium subspecies hominissuis (MAH) causes disease in both humans and swine. We elucidated the genetic variations in MAH isolates from humans and swine in Japan. We analyzed the 16S-23S rDNA internal transcribed spacer (ITS) sequence and variable number of tandem repeats (VNTR) using the Mycobacterium avium tandem repeat (MATR) loci, prevalence of ISMav6 and clarithromycin (CLA) resistance for MAH isolates from patients with pulmonary MAC (pMAC) disease (n = 69), HIV-seropositive and blood culture-positive (HIV-MAC) patients (n = 28), and swine (n = 23). In the minimum spanning tree (MST) based on VNTR analysis, swine MAC isolates belonged to a cluster distinguishable from that of human pMAC isolates. Isolates from HIV-MAC were scattered throughout both clusters. The major three distinct sequevars, Mav-A, B, F were determined according to 16S-23S rDNA ITS sequence analysis in addition to three new sequevars, Mav-Q, R, and S. Mav-A and Mav-F comprised the majority of human pMAC strains, in contrast, Mav-B was predominated in swine isolates. Distribution of ITS sequevars in the MST based on VNTR analysis showed similar clusters of isolates from different origins i.e. human pMAC, HIV-MAC and swine. These results, together with ISMav6 possession and CLA resistance, showed the distinctive genetic evolutionary lineage of MAH strains isolated from human pMAC diseases and swine.
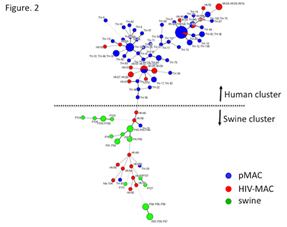
VNTR
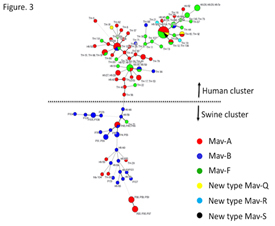
16S-23S rDNA ITS sequence
Faculty Members
| Faculty | Position | Department |
|---|---|---|
| Tetsuya Yagi | Professor | Department of infectious diseases |
| Yuka Tomita | Assistant professor | Department of infectious diseases |
| Daizo Kato | Assistant professor | Department of infectious diseases |
| Mitsutaka Iguchi | Assistant professor |
Bibliography
- 2016
- Adachi T, Ichikawa K, Inagaki T, Moriyama M, Nakagawa T, Ogawa K, Hasegawa Y, Yagi T. Molecular typing and genetic characterization of Mycobacterium avium subspecies hominissuis isolates from humans and swine in Japan. J Med Microbiol 2016 Sep 6. Doi: 10.1099/jmm.0.000351.
- Morioka H, Hirabayashi A, Iguchi M, Tomita Y, Kato D, Sato N, Hyodo M, Kawamura N, Sadomoto T, Ichikawa K, Inagaki T, Kato Y, Kouyama Y, Ito Y, Yagi T. The first point prevalence survey of healthcare-associated infection and antimicrobial use in a Japanese university hospital: A pilot study. Am J Infect Control , 2016;44: 119-23
- 2015
- Okachi S, Wakahara K, Kato D, Umeyama T, Yagi T, Hasegawa Y. Massive mediastinal cryptococcosis in a young immunocompetent patient. Respirol Case Rep , 2015;3: 95-8.
- Ichikawa K, van Ingen J, Koh WJ, Wagner D, Salfinger M, Inagaki T, Uchiya K, Nakagawa T, Ogawa K, Yamada K, Yagi T.;Genetic diversity of clinical Mycobacterium avium subsp. hominissuis and Mycobacterium intracellulare isolates causing pulmonary diseases recovered from different geographical regions. Infection, Genetics and Evolution, 2015;36: 250-255.
- Shindo Y, Ito R, Kobayashi D, Ando M, Ichikawa M, Goto Y, Fukui Y, Iwaki M, Okumura J, Yamaguchi I, Yagi T, Tanikawa Y, Sugino Y, Shindoh J, Ogasawara T, Nomura F, Saka H15, Yamamoto M, Taniguchi H, Suzuki R, Saito H, Kawamura T, Hasegawa Y. Central Japan Lung Study Group. Risk factors for 30-day mortality in patients with pneumonia who receive appropriate initial antibiotics: an observational cohort study. Lancet Infect Dis, 2015 ;15:1055-65.
- Uchiya K, Takahashi H, Nakagawa T, Yagi T, Moriyama M, Inagaki T, Ichikawa K, Nikai T, Ogawa K. Characterization lf a Novel plasmid,pMAH135,from Mycobacterium avium subsp.hominissuis. PLOS ONE, 2015;10:e0117797.
- Asai K, Yamada K, Yagi T, Baba H, Kawamura I, Ohta M. Effect of incubation atmosphere on the production and composition of staphylococcal biofilms. J Infect Chemother , 2015;21:55-61.
- Ito R, Shindo Y, Kobayashi D, Ando M, Jin W, Wachino JI, Yamada K, Kimura K, Yagi T, Hasegawa Y, Arakawa Y. Molecular epidemiological characteristics of Klebsiella pneumoniae associated with bacteremia among patients with pneumonia. J Clin Microbiol , 2015;pii:JCM.03067-14.
- 2014
- Nakamura G, Wachino JI, Sato N, Kimura K, Yamada K, Jin W, Shibayama K, Yagi T, Kawamura K, Arakawa Y. Practical agar-based disk potentiation test for detection of fosfomycin nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J Clin Microbiol , 2014;52:3175-3179.
- 2013
- Shindo Y, Ito R, Kobayashi D, Ando M, Ichikawa M, Shiraki A, Goto Y, Fukui Y, Iwaki M, Okamura J, Yamaguchi I, Yagi T, Tanikawa Y, Sugino Y, Shindou J, Ogasawara T, Nomura F, Saka H, Yamamoto M, Taniguchi H, Suzuki R, Saito H, Kawamura T, Hasagawa Y. Risk factors for drug-resistant pathogens in community-acquired and healthcare-associated pneumonia. Am J Respir Crit Care Med , 2013;188:985-995.
- Yamaguchi M, Terao Y, Mori-Yamaguchi, Domon H, Sakaue Y, Yagi T, Nishino K, Yamaguchi A, Nizet V, Kawabata S. Streptococcus pneumoniae invades erythrocytes and utilizes them to evade human innate immunity. PLOS ONE, 2013;8:e77282.
- Uchiya K, Takahashi H, Yagi T, Moriyama M, Inagaki T, Ichikawa K, Nakagawa T, Nikai T, Ogawa K. Comparative genome analysis of Mycobacterium avium revealed genetic diversity in strains that cause pulmonary and disseminated disease. PLOS ONE, 2013 ;8:e71831
- Matsumura Y, Nagao M, Iguchi M, Yagi T, Komori T, Fujita N, Yamamoto M, Matsushima A, Takakura S, Ichiyama S.Molecular and clinical characterization of plasmid-mediated AmpC β-lactamase-producing Escherichia coli bacteraemia: a comparison with extended-spectrum β-lactamase-producing and non-resistant E. coli bacteraemia. Clin Microbiol Infect , 2013 ;19:161-168.
Research Keywords
Infection prevention and control、 healthcare-associated infections、 drug resistant bacteria、 antimicrobial therapy、 non-tuberculous mycobacteria
Call for graduate students
We’re always open to young doctors who are interested in prevention, control and treatment of infectious diseases.
Come and join us!!