FexSplice predicts a splicing consequence of a single nucleotide variation (SNV) at the first nucleotide of an exon (Fex-SNV). Fex-SNV affects splicing at AG-dependent 3’ splice sites (ss), in which the polypyrimidine tract (PPT) is short or degenerate, and requires binding of U2AF35 at the intron/exon boundary to reinforced the binding of U2AF65 to PPT. In contrast, Fex-SNV does not affect splicing at AG-independent 3’ ss, in which PPT is long. When the first nucleotide of an exon is not G, binding of U2AF35 to the intron-exon boundary becomes weak, and such 3’ ss’s are mostly AG-independent.
FexSplice was generated by LightGBM with 106 splicing-affecting and 106 neutral Fex-SNVs extracted from Human Gene Mutation Database Professional (HGMD Pro) released in April 2020 and ClinVar with CLNSIG = pathogenic released in March 2021, and from dbSNP build 151 on GRCh37/hg19 with minor allelic frequency (MAF) greater than 0.01 and less than 0.5, respectively. A total of 115 features dictating the strength of splicing cis-elements, which included the length and nucleotide composition of the polypyrimidine tract; the composition of the branch point sequence; the nucleotides at intronic positions -6 and -5 and exonic positions +2 and +3 that are critical determinants of splicing in our previous analysis (Shibata A. et al. J Hum Genet 2016, 61, 633-640, doi:10.1038/jhg.2016.23), MaxEntScan scores at the 3' and 5' splice sites (Yeo G. et al. J Comput Biol 2004, 11, 377-394, doi:10.1089/1066527041410418), and the presence of recognition motifs of each RBP according to SpliceAid2 (Piva F. et al. Hum Mutat 2012, 33, 81-85, doi:10.1002/humu.21609). Recursive elimination of features by cross-validation showed that 15 features best predicted the splicing consequences of Fex-SNVs with an area under the curve of the receiver operating characteristics curve of 0.86 ± 0.08 (mean and SD) by 10-fold cross-validation.
LightGBM automatically generates a probability score for each Fex-SNV with 0.5 being the threshold. The default threshold of 0.5 by LightGBM was used in FexSplice. Fex-SNVs with a probability less than 0.5 are predicted to be splicing-insensitive, while those with a probability of 0.5 or more are predicted to be splicing-affecting.
Citation:
-
Joudaki A, Takeda J, Masuda A, Ode R, Fujiwara K, Ohno K. FexSplice: a LightGBM-based model for predicting the splicing effect of a single nucleotide variant affecting the first nucleotide G of an exon. Genes. 14(9): 1765, 2023. (PMID: 37761905)
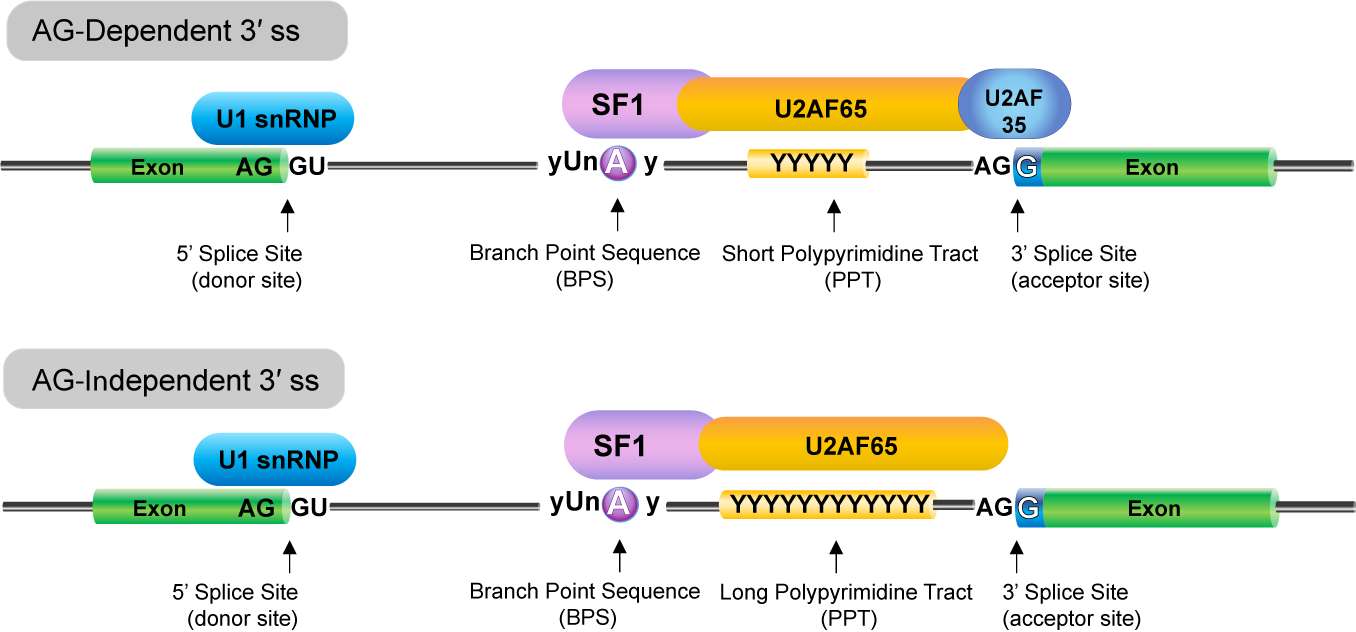
Figure 1. AG-dependent and AG-independent 3’ splice sites (ss). Introns with a short or degenerate PPT require both U2AF65 and U2AF35 for the recognition of the 3′ ss, which is called the AG‐dependent 3′ ss. Introns with a long stretch of PPT strongly binds to U2AF65 and does not require binding of U2AF35, which is called the AG‐independent 3′ ss. The 3’ ss’s without G at the first nucleotide of an exon are mostly AG‐independent.